Scientists at Harvard Medical School have made freely available an artificial intelligence tool, PDGrapher, that can look for drug targets capable of steering diseased cells back to health-happy behavior. Preliminary results suggest it could speed treatments for stubborn illnesses, from multiple cancers to neurodegenerative diseases like Parkinson’s and Alzheimer’s.
How Harvard’s PDGrapher creates maps of disease networks
PDGrapher calculates how genes, proteins and signaling pathways interact within cells and determines which levers — singly or in combination — are most likely to return a disease state to normal physiological operation.
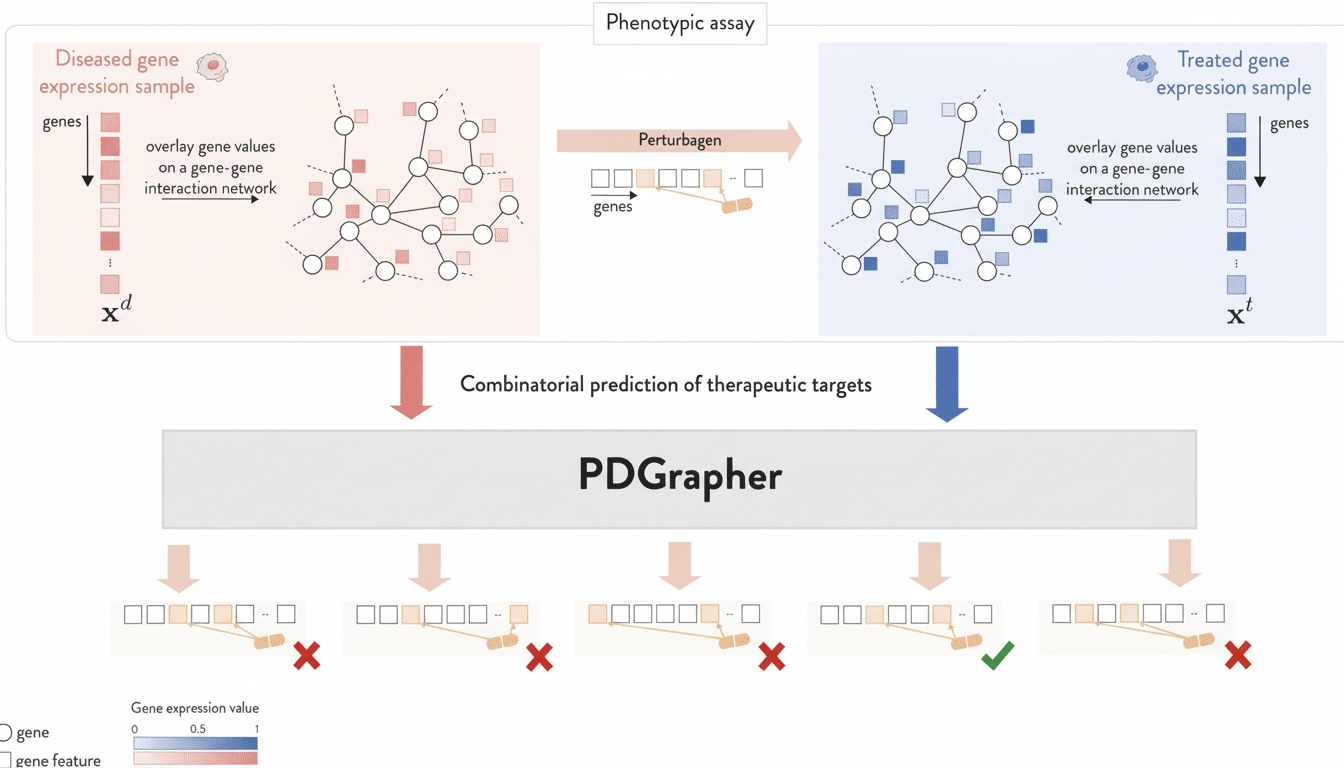
Rather than evaluating one protein target at a time, the model learns from large numbers of cell profiles taken before and after treatments to determine where interventions are most likely to have the biggest impact.
The method is based on network biology. Many cancers and brain diseases have twisted signaling cascades at their core, rather than a single busted protein. By empowering users to treat the cell as a coherent system, PDGrapher has been able to bring up multi-drug approaches that may otherwise be missed by classical protocols Students describe it as moving from “trial-and-error tasting” to the designing of a new recipe for healthful function.
Benchmarked performance on cancer datasets
To see how well the approach could generalize, the researchers tested PDGrapher on 19 data sets from 11 different cancer types and tasked it with predicting effective therapeutic targets in cell samples it hadn’t seen before. The tool correctly ranked clinically validated targets and discovered new putative targets that had support available in the literature.
And in head-to-heads, the researchers found that PDGrapher ranked correct targets 35% higher than competing methods — and did so 25 times faster.
That blend of accuracy and speed counts in drug discovery, where getting moderately months earlier to target selection can free resources from dead ends up into promising biology.
Why multi-target strategies matter
Cancer care for some patient subsets has been transformed by single-target drugs — many kinase inhibitors, among other targeted agents. But complex diseases would bypass a blocked node all the time. The presence of resistance in oncology and the multifactorial character of neurodegeneration make clear why hitting more than one pathway can be crucial.
In Alzheimer’s, for instance, pathology runs the gamut from protein aggregation to synaptic failure to neuroinflammation. Studies cited by the Alzheimer’s Association have found extremely high failure rates for experimental drugs over the past 20 years. Parkinson’s disease is also a complex web of events, from mitochondrial dysfunction to alpha-synuclein toxicity. A system-level tool that offers possible combinations or sequence of targets based on systematically known biological constraints could facilitate the design of interventions more resilient to disease networks escape.
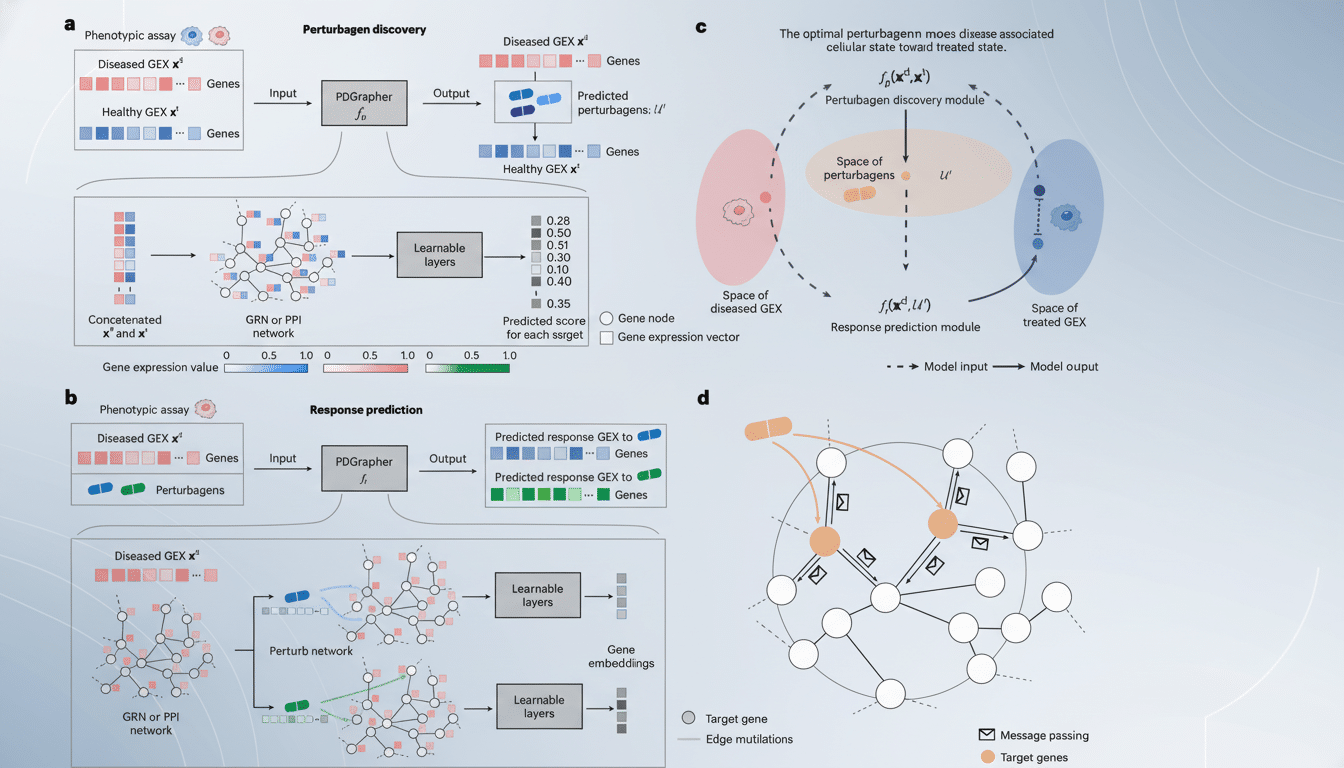
What this means for Parkinson’s and Alzheimer’s research
The Harvard team is already using PDGrapher in the context of brain disorders, where living tissue cannot be easily accessed and mechanistic uncertainty is high. By sifting through perturbation data and disease signatures, the model can spotlight convergent pathways — say, immune signaling combined with proteostasis — that collectively conspire to cause neuronal dysfunction, and then nominate target sets to test in cellular or animal models.
That ability paves the way for repurposed drugs, rational combination design and biomarker-guided trials.” It might also guide decisions about which targets to go after with gene therapies or antisense strategies, inroads that are getting more attention in neurology.
Open, free and designed for collaboration
PDGrapher is publicly available to other researchers from the authors’ public code repository, representing a step toward open and reproducible AI in biomedicine. The research was partly federally funded, and its authors stress that their tool is not to replace, but rather to supplement laboratory validation.
Being open means academic labs and biotech teams can plug in their own disease datasets, retrain a model to a specific context, and benchmark against internal pipelines. That kind of interoperability might enable smaller groups to take advantage of cutting-edge target discovery without building bespoke models from the ground up.
Caveats and what to watch next
Like all such models, PDGrapher’s predictions are only as good as the data on which it trains. Most training references are from in vitro systems, and conversion to human biology has required careful validation; in need of functional assays, clinical studies. There is also a constant danger of confounding if some cell types or stages of disease are underrepresented in training sets.
Even with those caveats, the early output from PDGrapher suggests a practical advance: a swift, clear engine for transforming complex molecular maps into testable therapeutic hypotheses.
If continuing work in oncology, Parkinson’s and Alzheimer’s backs those gains up, the tool could help shorten the route from target to treatment.

